Regions
Regions are used to isolate spectral ranges for certain operations. There are three different types of regions:
- Continuum: used for fitting the (pseudo-)continuum (instead of using the whole spectrum).
- Line masks: used for gaussian fitting (e.g. equivalent width measurements) and/or atmospheric parameters/abundance determination. In the latter case, only the fluxes from the spectral ranges inside line masks are going to be considered for the \(\chi^2\) minimization.
- Segments: used mainly for atmospheric parameters/abundance determination. The synthetic spectra is going to be computed only for the spectral ranges inside segments, thus they should include all the line masks (grouped in one or more segments). Segments permit to save computation time, avoiding to compute the whole synthetic spectra, and they should be larger than the line masks (e.g., if a line with a line mask is in a blended region, such as close to H-\(\alpha\), the segment should be big enough to cover the blended region or the synthesis might not well reproduce that spectral range).
For creating, modifying or removing regions, an action and an element should be selected in iSpec:
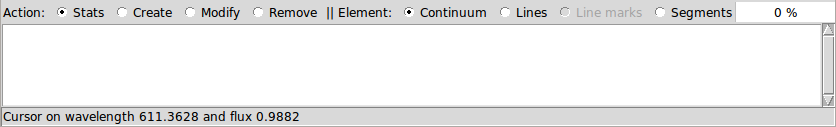
The possibilities with each combination are explained in the following table (it is worth noting that the mouse position in wavelength/flux can be found in the status bar of the editor's window):
| Stats | Create | Modify | Remove | |
|---|---|---|---|---|
| Continuum / Lines / Segments | Left click on a region and its statistical information will be visible in the bottom part of the window. | Left click on an empty space and a new region will appear. If it is a line region, it will ask for an optional note. | Left click and drag modifies the left edge. Right click and drag modifies the right edge. | Left click to remove the region. |
| Line marks | - | Left click on a line region to add a note. | Left click on a line region to modify the peak mark. Right click on a line region to add/modify a note. | Left click on a line region to remove the note. |
It is important to remark that Zoom/Pan mode should be disabled in order to be able to execute the above actions. On the other hand, the user is responsible for not creating incoherent overlapping regions like line masks that overlap or segments without line masks.
Region files
Regions can be saved and read through the menu "File". Once loaded, they can also be completely removed from the plot through the menu "Operations - Clear - Continuum/Line masks/Segments". In case some regions have been modified and not saved, iSpec will ask for confirmation before clearing them.
iSpec can also be executed from command line indicating directly the regions and spectra to be opened:
./iSpec.command --continuum=input/regions/fe_lines_continuum.txt \
--lines=input/regions/fe_lines.txt \
--segments=input/regions/fe_lines_segments.txt \
input/spectra/examples/NARVAL_Sun_Vesta-1.txt.gzContinuum regions file format
Continuum region files should be plain text files with tab character as column delimiter. Two columns should exists: 'wave_base' and 'wave_top' (the first line should contain those header names). They indicate the beginning and end of each region (one per line). For instance:
wave_base wave_top
480.6000 480.6100
481.1570 481.1670
491.2240 491.2260
492.5800 492.5990Additionally, "wave base" should be always lower than "wave top" or iSpec will not be able to process the file.
Line regions file format
Line region files should be plain text files with tab character as column delimiter. Four columns should exists: 'wave_peak', 'wave_base', 'wave_top' and 'note' (the first line should contain those header names). They indicate the peak of the line, beginning and end of each region (one per line) and a note (it can be any string comment). For example:
wave_peak wave_base wave_top note
480.8148 480.7970 480.8330 Fe 1
496.2572 496.2400 496.2820 Fe 1
499.2785 499.2610 499.2950
505.8498 505.8348 505.8660 Fe 1The note can be blank but the previous tab character should exists anyway.
Regarding the wavelenghts, "wave base" should be always lower than "wave top" and "wave peak" should be in between or iSpec will not be able to process the file.
Segment regions file format
Segment region files should be plain text files with tab character as column delimiter. Two columns should exists: 'wave_base' and 'wave_top' (the first line should contain those header names). They indicate the beginning and end of each region (one per line). For instance:
wave_base wave_top
480.6000 480.6100
481.1570 481.1670
491.2240 491.2260
492.5800 492.5990The values in the column "wave base" should be always lower than "wave top" or iSpec will not be able to process the file.
Automatic continuum regions
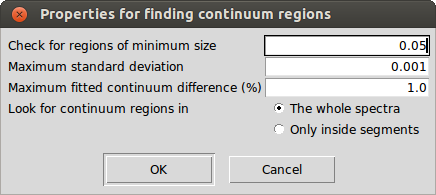
It is possible to find continuum regions automatically by analyzing the spectra slice by slice. A spectral range is selected as continuum if the following conditions are met:
- The region is at least as big as the size specified by the user.
- The standard deviation specified by the user is less than a given maximum.
- The median flux is above or below the continuum fit but not more than a given percentage.
Depending on the spectrum type, the slice size and the standard deviation can be adjusted to find better results.
This functionality is located in the "Operations - Find continuum regions" menu and can be applied over the whole spectra or only inside the defined segments. In both cases, it needs a previously fitted continuum. After the computation, it removes the current continuum regions (if there are any) and draws the ones that the process has found.
Automatic line masks
iSpec can find line regions automatically by selecting "Operations - Find line masks". The search can be done over the whole spectra or only inside the defined segments. In both cases, it needs a previously fitted continuum.
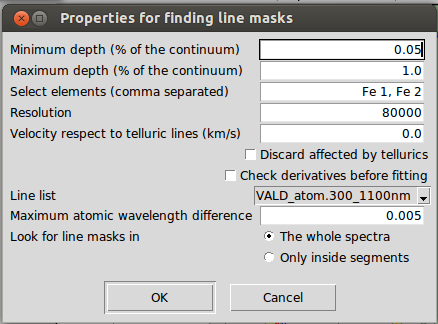
The process follows these steps:
- Search local maximum and minimum points of a spectrum that has been smooth by using two times the resolution.
- Select those line candidates that have a minimum depth (1.0 represents 100\% of depth with respect to the continuum).
- Fit the line candidates with a Gaussian model and discard those with bad fit. If "check derivatives" is active, the first and second derivatives of the line candidates are checked to try to better delimit where it begins and end (this works better for synthetic spectra or observed spectra with very high signal-to-noise ratios).
- Cross-match the remaining lines with an atomic line list considering the velocity specified by the user. It is recommended to have corrected the radial velocity previously, so that the velocity here would be zero.
- Select the lines that correspond to the elements specified by the user (comma separated or blank to avoid this filter).
- Cross-match again with a telluric line list considering the velocity specified by the user. Again, it is recommended to have corrected the radial velocity previously.
- Discard lines that may be affected by telluric lines if the user has indicated so.
After the computation, it removes the current line regions (if there are any) and draws the ones that the process has found. Note that the cross-match with the atomic data is not always 100% true, since there can be observed absorption lines for which there is no information on the atomic list, or blended lines which the main contributor is not the one it has cross-matched from the atomic line list, etc.
For atmospheric parameters or abundance determination, it is recommended to use a selection of lines that have been shown to work correctly for this kind of task. Not all the lines are good enough to derive parameters. iSpec includes line selections that will be discussed in the section about parameter and abudance determination.
Adjust line masks
Line masks automatically found or manually defined for a given type of stars may not fit well enough the shape of the line. iSpec can adjust automatically previous defined line masks to match a better beginning/end of the masks to the particular form of the active spectrum. If "check derivatives" is active, the first and second derivatives of the line candidates are checked to try to better delimit where it begins and end (this works better for synthetic spectra or observed spectra with very high signal-to-noise ratios).
To do so, iSpec will check were is the optimal limit of the line by looking at a window of a given margin (in nanometers) around the line center. The algorithm will search for the local maximum in each side of the line that is closer to the line center.
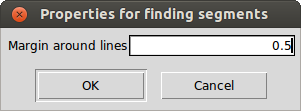
Create segments around line masks
The user that has a group of line masks already defined could be interested in creating segments around them, for instance, in order to compute synthetic spectra or derive atmospheric parameters. iSpec can do that automatically, the user should specify how many nanometers he wants around each line masks and iSpec will group those lines that are close enough under the same segment.